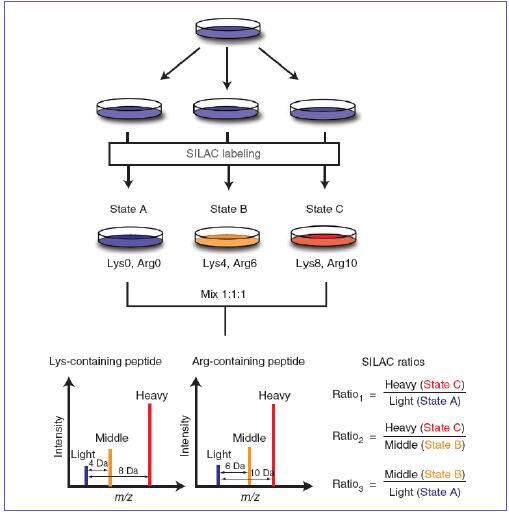
Multiple studies with a single experiment: The Power of Quantitative Multiplexing
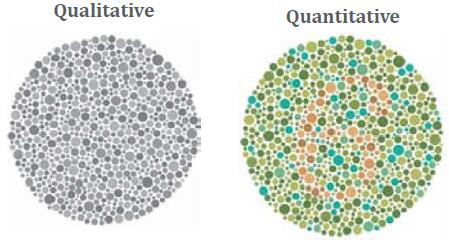



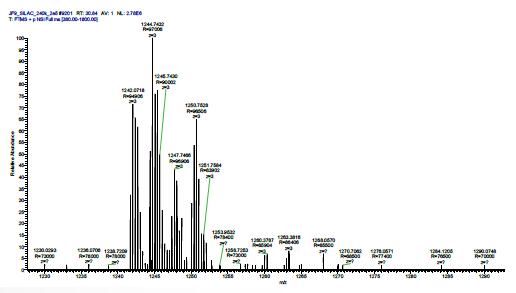

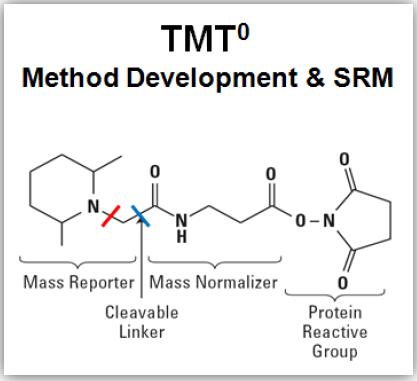
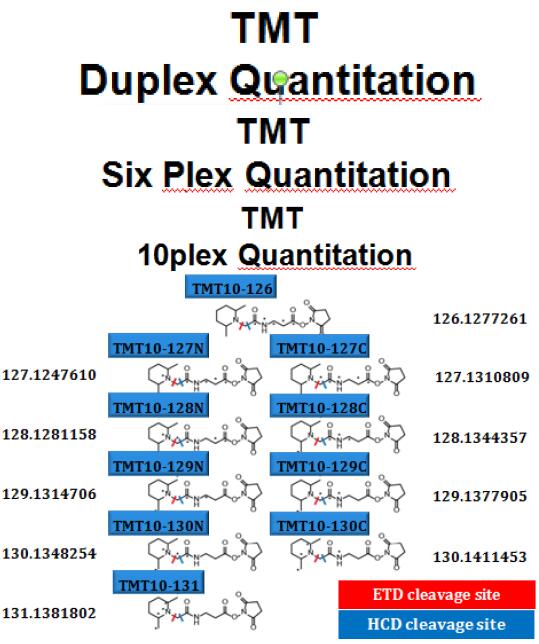
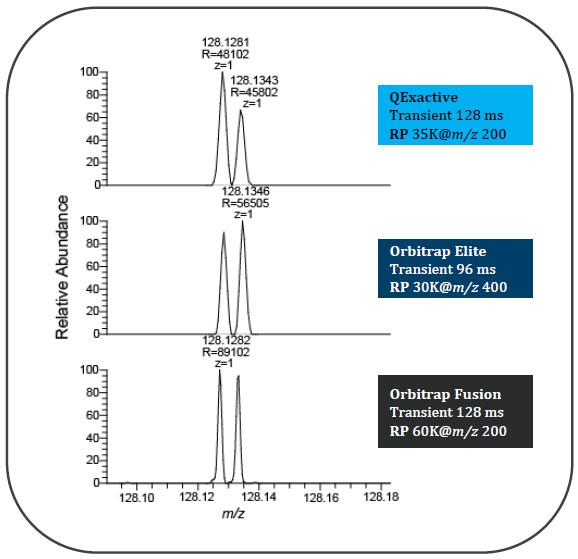
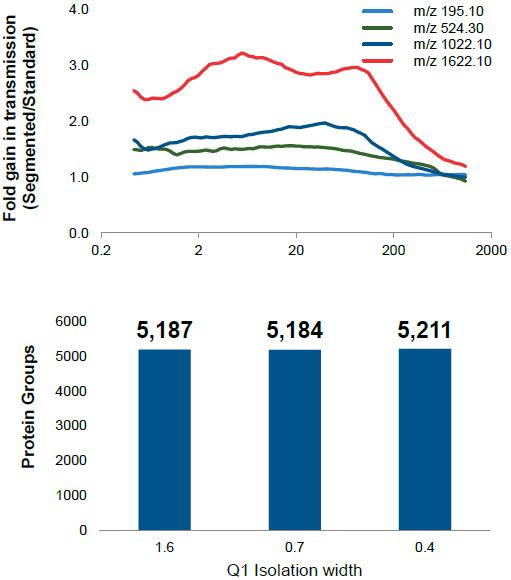
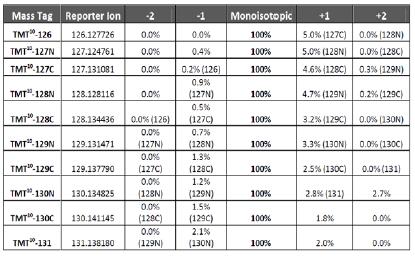
HIGH RESOLVING POWER IS ESSENTIAL FOR ACCURATE QUANTITATION OF THE TMT10PLEX REAGENTS
Result: Get accurate quantitation using the high resolution of Orbitrap Mass Analyzer
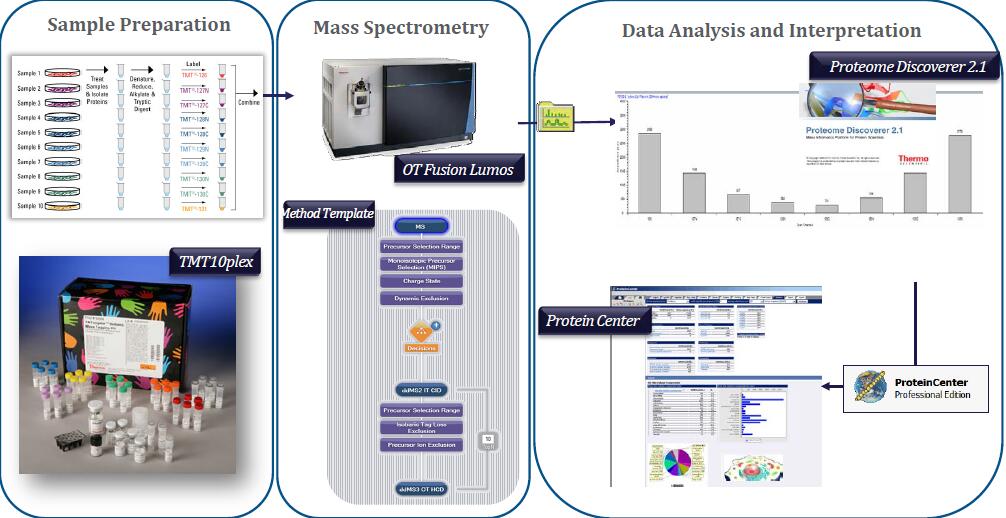
A Real Example
Sample: Mouse mitochondrial extract untreated or treated with phosphatase inhibitor
Orbitrap Elite
*75 um x 50 cm PepMap C18
*210 min gradient: 250 min run
*1 ug of sample on column

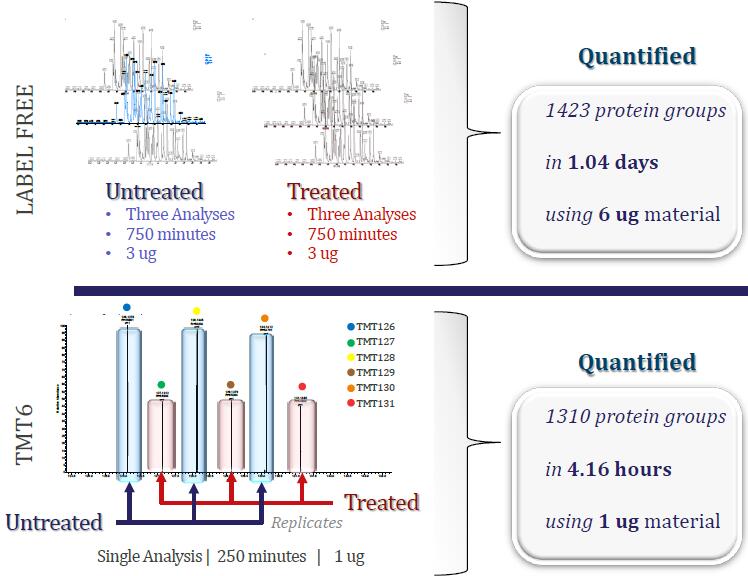
Thermo Poster Note : Liver Mitochondria Proteomics Employing High –Resolution MS Technology; J.Ho. et al
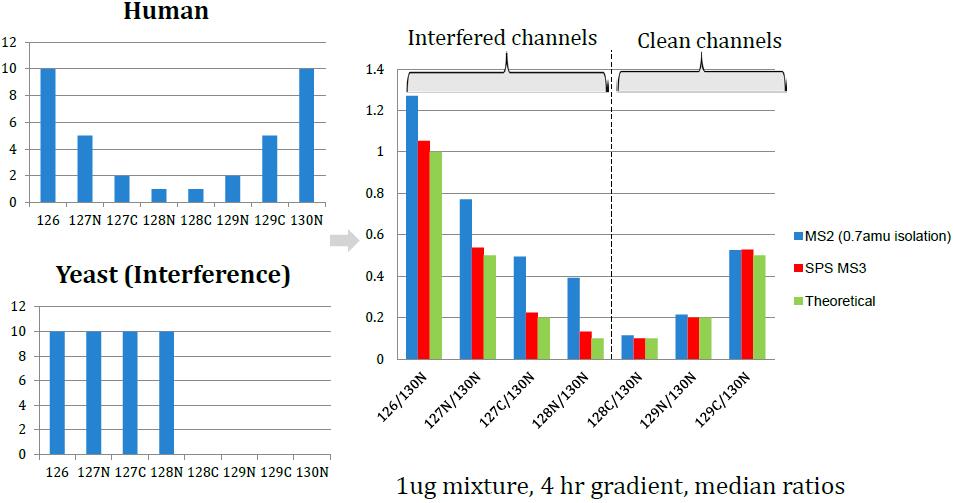
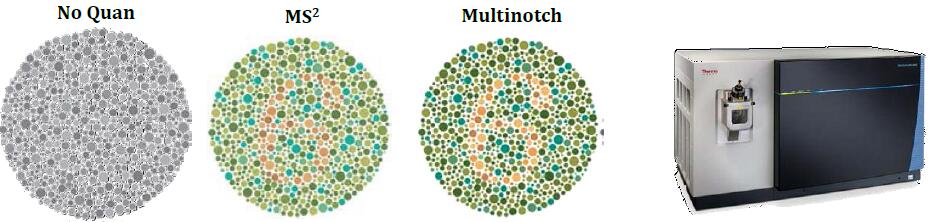
Ratio Distortion with Isobaric Multiplexing
Problem: Quantitation of low-abundance proteins in a complex background is distorted by co-isolated interfering precursor ions

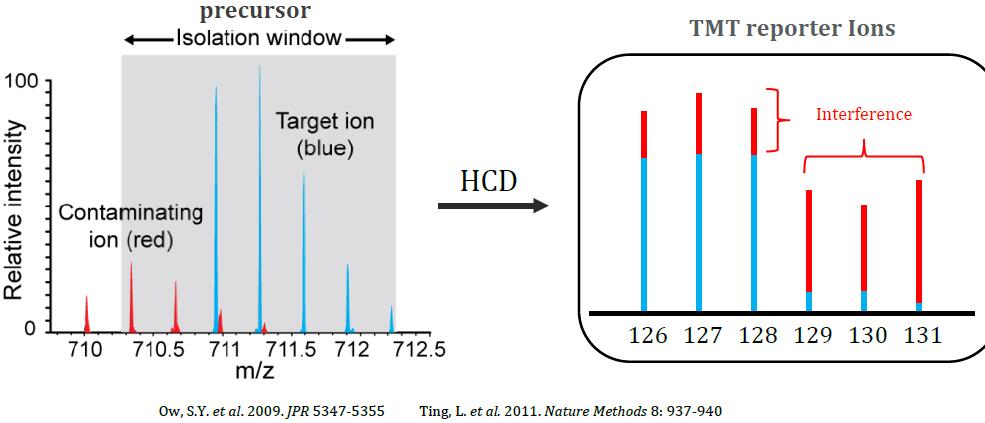
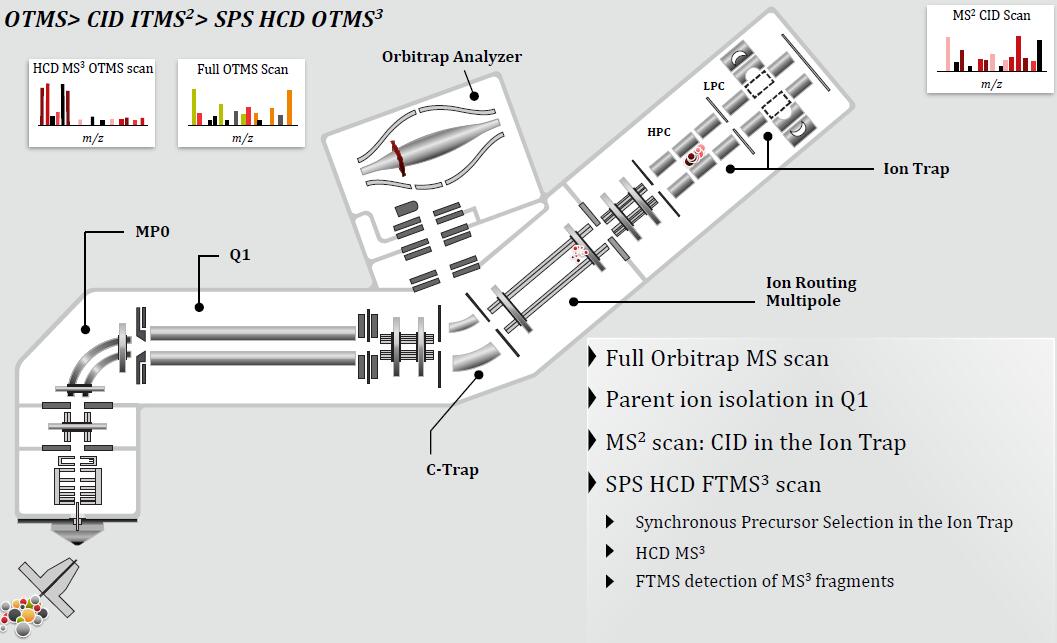
Synchronous Precursor Selection (SPS) for Accurate Quantification
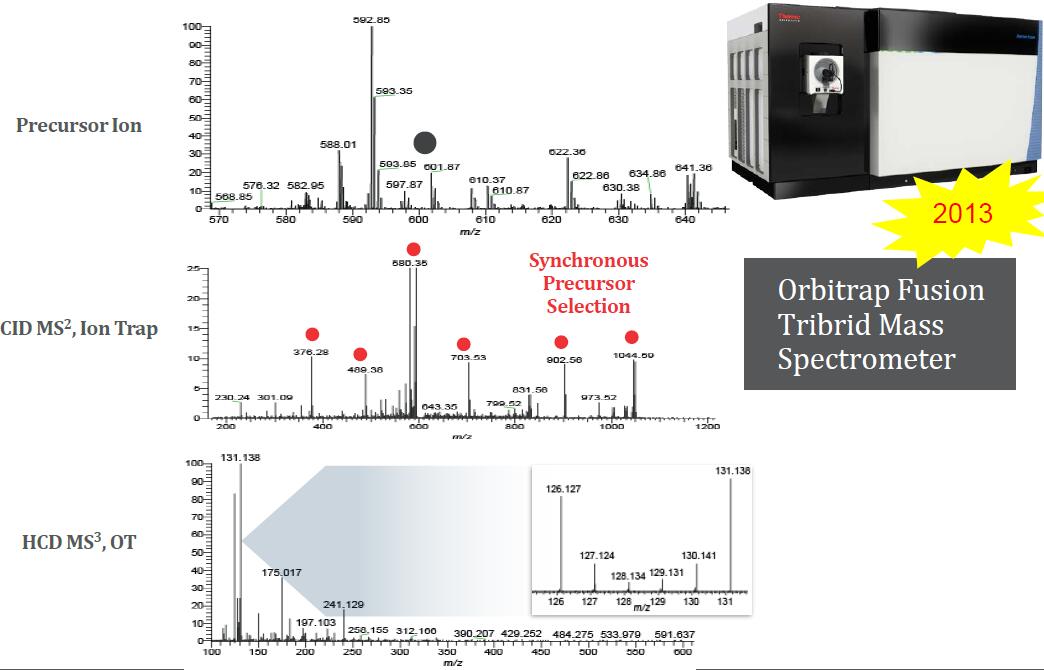
TMT3 Experiment, Powered by SPS
Co-isolation of Interfering Ions Affects Accuracy
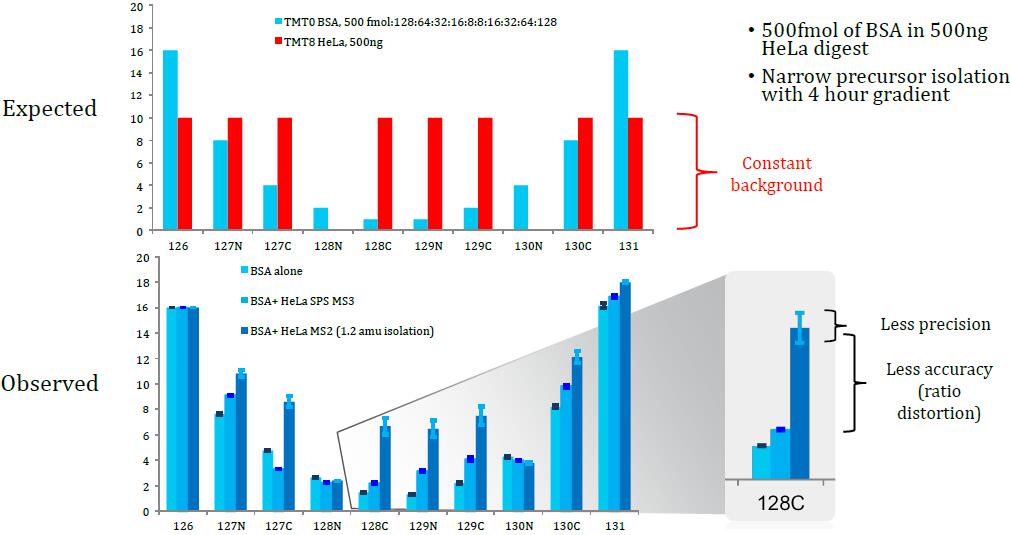
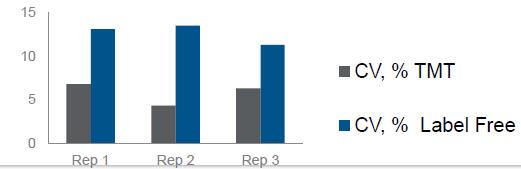
Results: Best possible accuracy and precision by reducing co-isolated interfering ions.
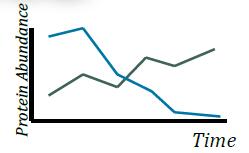
Enhanced Differences Using SPS MS3 Quantitation
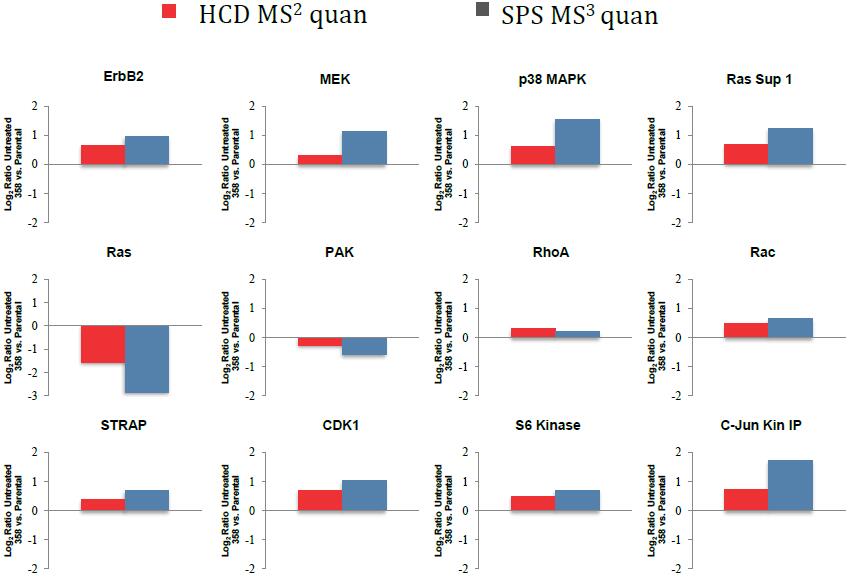
Thermo Poster Note : Towards Mechanism of EGFR Inhibitor Resistance in Non-Small Lung Cancer Cell;M.Blank. et al
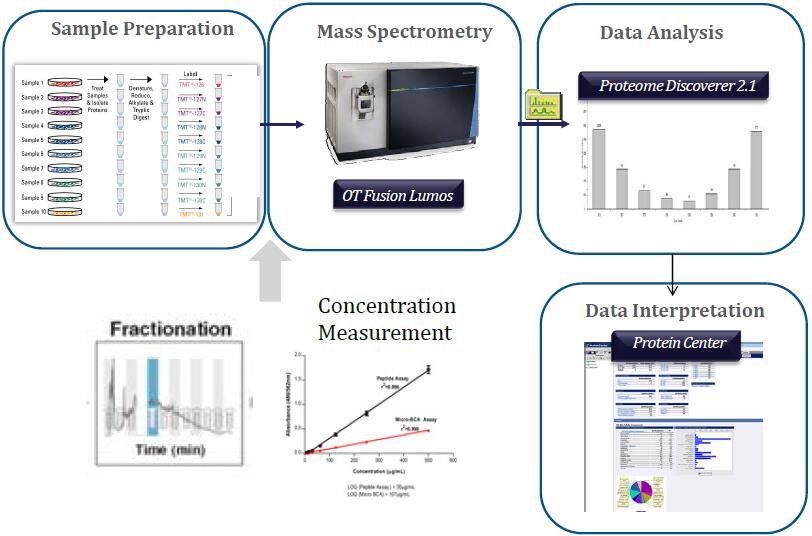
…While Still Getting Proteome Coverage
The speed and parallelizable work flow of the Orbitrap Fusion means not choosing between accuracy and coverage…
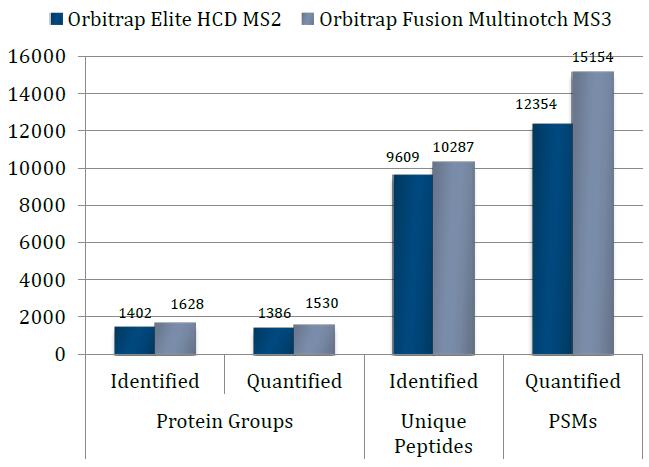
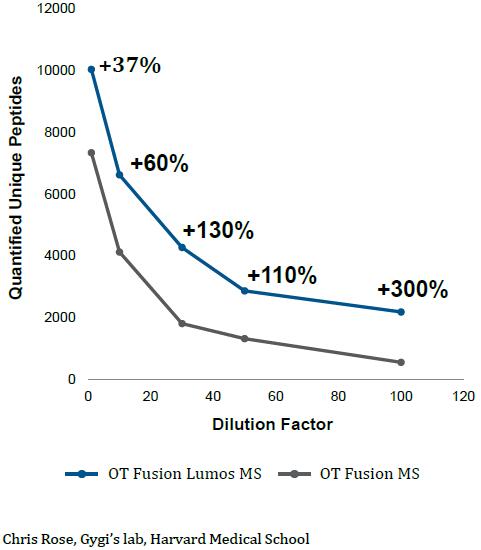
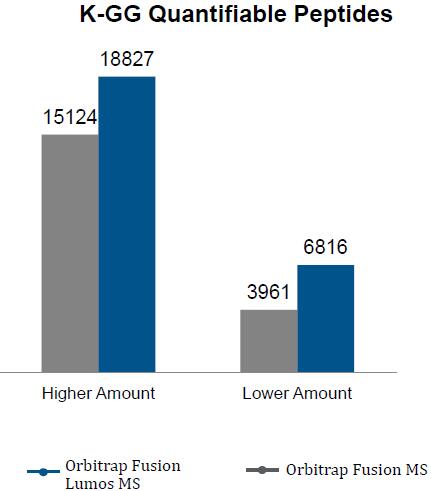
The Orbitrap Fusion using multinotch can Quantify more proteins than were Identified on the previous generation top tier hybrid
…Don’t Settle
Orbitrap Fusion Lumos Tribrid Mass Spectrometer
2015

Unmatched Analytical Performance
Revolutionary performance
Exceptional versatility
Unprecedented usability
Highest sensitivity








•Ryan Bomgarden
•Sergei Snovida
•Paul Haney
•John Rogers
•Rosa Viner
•Xiaoyue Jiang
•Michael Blank
•Andreas Huhmer
•Graeme McAllister
•Tabiwang Arrey
•Vlad Zabrouskov
•Michaela Scigelova
•David Horn
•Torsten Ueckert
•Steve Gygi, Harvard Medical School
•Josh Coon, University of Wisconsin, Madison
•Jennifer Van Eyk, Johns Hopkins School of Medicine
•Zezong Gu, University of Missouri-Columbia
•Kay-Hooi Khoo, Academia Sinica
•Bernhard Kuster, Technische Universität München
•Somi Afiuni & Tim Griffin, University of Minnesota